# Run this cell to install the latest version of fastai2 shared on github
!pip install git+https://github.com/fastai/fastai2.git
# Run this cell to install the latest version of fastcore shared on github
!pip install git+https://github.com/fastai/fastcore.git
# Run this cell to install the latest version of timeseries shared on github
!pip install git+https://github.com/ai-fast-track/timeseries.git
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai2.basics import *
from timeseries.all import *
Class Activation Map (CAM) and Grafient-CAM (GRAD-CAM) Tutorial
Both CAM and GRAD-CAM allow producing ‘visual explanations’ on how a Convolutional Neural Network (CNN) model based its classification and therefore help interpreting the obtained results. The InceptionTime model is used as an illustration in this notebook.
ECG Dataset
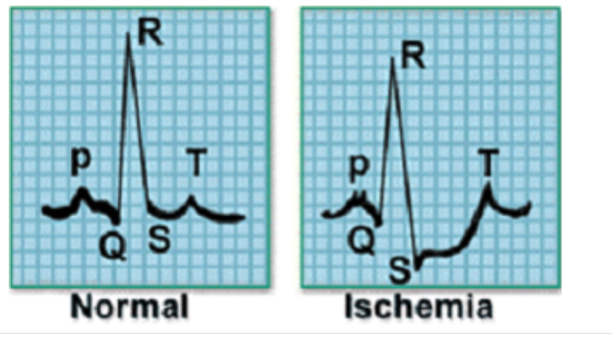
This dataset was formatted by R. Olszewski as part of his thesis “Generalized feature extraction for structural pattern recognition in time-series data,” at Carnegie Mellon University, 2001. Each series traces the electrical activity recorded during one heartbeat. The two classes are a normal heartbeat and a Myocardial Infarction. Cardiac ischemia refers to lack of blood flow and oxygen to the heart muscle. If ischemia is severe or lasts too long, it can cause a heart attack (myocardial infarction) and can lead to heart tissue death.
# You can choose any of univariate dataset listed the `data.py` file
path = unzip_data(URLs_TS.UNI_ECG200)
dsname = 'ECG200' # 'GunPoint'
fname_train = f'{dsname}_TRAIN.arff'
fname_test = f'{dsname}_TEST.arff'
fnames = [path/fname_train, path/fname_test]
fnames
# num_workers=0 is for Windows platform
dls = TSDataLoaders.from_files(bs=64,fnames=fnames, num_workers=0)
dls.show_batch(chs=range(0,12,3))
learn = ts_learner(dls)
learn.fit_one_cycle(25, lr_max=1e-3)
learn.show_results()
model = learn.model.eval()
model[5]
dls.vocab
# i2o() function
# Converting CategoryTensor label into the human-readable label
lbl_dict = dict([
(0, 'Normal'),
(1, 'Myocardial Infarction')]
)
def i2o(y):
return lbl_dict.__getitem__(y.data.item())
# return lbl_dict.__getitem__(int(dls.tfms[1][1].decodes(y)))
idxs = [0,2]
batch = get_batch(dls.train.dataset, idxs)
Class Activation Map (CAM)
This option calculates the activations values at the selected layer.By default the activations curves are plotted in one single figure.
func_cam=cam_acts : activation function name (activation values at the chosen model layer). It is the default value
The figure title [Myocardial Infarction - Normal] - CAM - mean should be read as follow:
Myocardial Infarction: class of the first curveNormal: class of the second curveCAM: activation function name (activation values at the chosen model layer)mean: type of reduction (read the explanation below: 4 types of reductions)
show_cam(batch, model, layer=5, i2o=i2o, func_cam=cam_acts) # default: func_cam=cam_acts, multi_fig=False, figsize=(6,4)
Using CAM and CMAP.seismic palette
show_cam(batch, model, layer=5, i2o=i2o, multi_fig=True, cmap=CMAP.seismic) # default: func_cam=cam_acts, figsize=(13,4)
Using GRAD-CAM and CMAP.seismic palette
Notice the difference in activations values between CAM and GRAD-CAM
show_cam(batch, model, layer=5, i2o=i2o, func_cam=grad_cam_acts, multi_fig=True, cmap=CMAP.seismic)
show_cam(batch, model, layer=5, i2o=i2o, func_cam=grad_cam_acts, cmap=CMAP.seismic)
show_cam(batch, model, layer=5, i2o=i2o, linewidth=2, scatter=True, cmap=CMAP.hot_r)
dls.train = dls.train.new(bs=5)
batch = dls.train.one_batch()
# batch
show_cam(batch, model, layer=5, i2o=i2o, cmap=CMAP.viridis)
show_cam(batch, model, layer=5, i2o=i2o, cmap=CMAP.viridis, multi_fig=True, figsize=(18, 8), linestyles='dotted')
show_cam(batch, model, layer=5, i2o=i2o, func_cam=grad_cam_acts, multi_fig=True, figsize=(18, 8))
Plotting CAM for a single dataset item
We can also feed the
show_cam()a single itemThere are also
164different palettes. Check outCMAPclass and its autocompletionline styles :'solid' | 'dashed' | 'dashdot' | 'dotted'
idxs = [0]
batch = get_batch(dls.train.dataset, idxs)
show_cam(batch, model, layer=5, i2o=i2o, cmap='rainbow', linewidth=2, linestyles='dotted')
