Method 1 : Editable Version
1A - Installing fastai2
Important :Only if you have not already installed
fastai2,install fastai2 by following the steps described there.
1B - Installing timeseries on a local machine
Note :Installing an editable version of a package means that you will install a package from its corresponding github repository on your local machine. By doing so, you can pull the latest version whenever a new version is pushed. To install
timeserieseditable package, follow the instructions here below:
git clone https://github.com/ai-fast-track/timeseries.git
cd timeseries
pip install -e .Method 2 : Non Editable version
Note :Everytime you run the
!pip install git+https:// ..., you are installing the package latest version stored on github. > Important :As both fastai2 andtimeseriesare still under development, this is an easy way to use them in Google Colab or any other online platform. You can also use it on your local machine.
# Run this cell to install the latest version of fastai shared on github
!pip install git+https://github.com/fastai/fastai2.git
# Run this cell to install the latest version of fastcore shared on github
!pip install git+https://github.com/fastai/fastcore.git
# Run this cell to install the latest version of timeseries shared on github
!pip install git+https://github.com/ai-fast-track/timeseries.git
%reload_ext autoreload
%autoreload 2
%matplotlib inline
from fastai2.basics import *
from timeseries.all import *
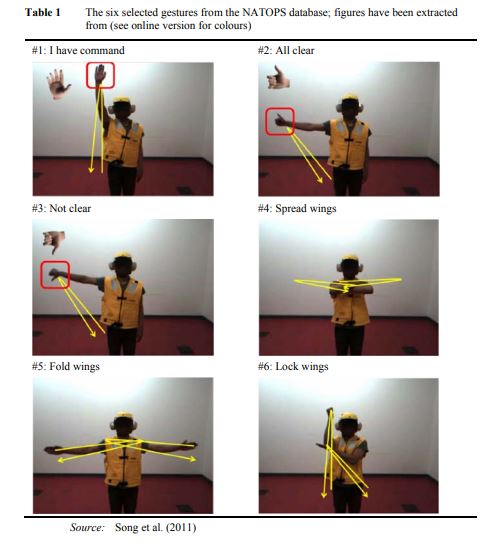
Right Arm vs Left Arm time series for the 'Not clear' Command ((#3) (see picture here above)
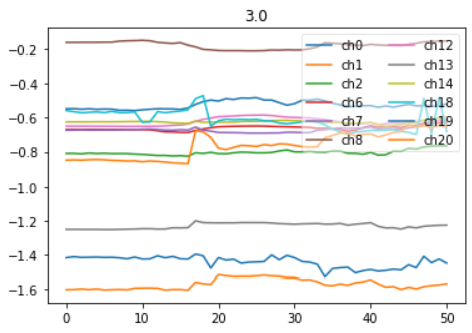
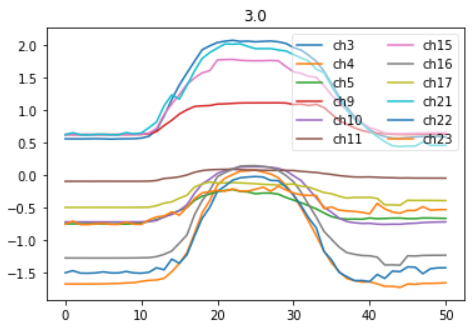
Channels (24)
| Hand | Elbow | Hand | Elbow |
|---|---|---|---|
| 0. Hand tip left, X | 6. Elbow left, X | 12. Wrist left, X | 18. Thumb left, X |
| 1. Hand tip left, Y | 7. Elbow left, Y | 13. Wrist left, X | 19. Thumb left, X |
| 2. Hand tip left, Z | 8. Elbow left, Z | 14. Wrist left, X | 20. Thumb left, X |
| 3. Hand tip righ, X | 9. Elbow righ, X | 15. Wrist righ, X | 21. Thumb righ, X |
| 4. Hand tip righ, Y | 10. Elbow righ, Y | 16. Wrist righ, X | 22. Thumb righ, X |
| 5. Hand tip righ, Z | 11. Elbow righ, Z | 17. Wrist righ, X | 23. Thumb righ, X |
dsname = 'NATOPS' #'NATOPS', 'LSST', 'Wine', 'Epilepsy', 'HandMovementDirection'
# url = 'http://www.timeseriesclassification.com/Downloads/NATOPS.zip'
path = unzip_data(URLs_TS.NATOPS)
path
fname_train = f'{dsname}_TRAIN.arff'
fname_test = f'{dsname}_TEST.arff'
fnames = [path/fname_train, path/fname_test]
fnames
data = TSData.from_arff(fnames)
print(data)
items = data.get_items()
idx = 1
x1, y1 = data.x[idx], data.y[idx]
y1
# You can select any channel to display buy supplying a list of channels and pass it to `chs` argument
# LEFT ARM
# show_timeseries(x1, title=y1, chs=[0,1,2,6,7,8,12,13,14,18,19,20])
# RIGHT ARM
# show_timeseries(x1, title=y1, chs=[3,4,5,9,10,11,15,16,17,21,22,23])
# ?show_timeseries(x1, title=y1, chs=range(0,24,3)) # Only the x axis coordinates
seed = 42
splits = RandomSplitter(seed=seed)(range_of(items)) #by default 80% for train split and 20% for valid split are chosen
splits
tfms = [[ItemGetter(0), ToTensorTS()], [ItemGetter(1), Categorize()]]
# Create a dataset
ds = Datasets(items, tfms, splits=splits)
ax = show_at(ds, 2, figsize=(1,1))
bs = 128
# Normalize at batch time
tfm_norm = Normalize(scale_subtype = 'per_sample_per_channel', scale_range=(0, 1)) # per_sample , per_sample_per_channel
# tfm_norm = Standardize(scale_subtype = 'per_sample')
batch_tfms = [tfm_norm]
dls1 = ds.dataloaders(bs=bs, val_bs=bs * 2, after_batch=batch_tfms, num_workers=0, device=default_device())
dls1.show_batch(max_n=9, chs=range(0,12,3))
getters = [ItemGetter(0), ItemGetter(1)]
tsdb = DataBlock(blocks=(TSBlock, CategoryBlock),
get_items=get_ts_items,
getters=getters,
splitter=RandomSplitter(seed=seed),
batch_tfms = batch_tfms)
tsdb.summary(fnames)
# num_workers=0 is Microsoft Windows
dls2 = tsdb.dataloaders(fnames, num_workers=0, device=default_device())
dls2.show_batch(max_n=9, chs=range(0,12,3))
getters = [ItemGetter(0), ItemGetter(1)]
tsdb = DataBlock(blocks=(TSBlock, CategoryBlock),
getters=getters,
splitter=RandomSplitter(seed=seed)
)
dls3 = tsdb.dataloaders(data.get_items(), batch_tfms=batch_tfms, num_workers=0, device=default_device())
dls3.show_batch(max_n=9, chs=range(0,12,3))
4th method : using TSDataLoaders class and TSDataLoaders.from_files())
dls4 = TSDataLoaders.from_files(fnames, batch_tfms=batch_tfms, num_workers=0, device=default_device())
dls4.show_batch(max_n=9, chs=range(0,12,3))
# Number of channels (i.e. dimensions in ARFF and TS files jargon)
c_in = get_n_channels(dls2.train) # data.n_channels
# Number of classes
c_out= dls2.c
c_in,c_out
model = inception_time(c_in, c_out).to(device=default_device())
model
# opt_func = partial(Adam, lr=3e-3, wd=0.01)
#Or use Ranger
def opt_func(p, lr=slice(3e-3)): return Lookahead(RAdam(p, lr=lr, mom=0.95, wd=0.01))
#Learner
loss_func = LabelSmoothingCrossEntropy()
learn = Learner(dls2, model, opt_func=opt_func, loss_func=loss_func, metrics=accuracy)
print(learn.summary())
lr_min, lr_steep = learn.lr_find()
lr_min, lr_steep
#lr_max=1e-3
epochs=30; lr_max=lr_steep; pct_start=.7; moms=(0.95,0.85,0.95); wd=1e-2
learn.fit_one_cycle(epochs, lr_max=lr_max, pct_start=pct_start, moms=moms, wd=wd)
# learn.fit_one_cycle(epochs=20, lr_max=lr_steep)
learn.recorder.plot_loss()
learn.show_results(max_n=9, chs=range(0,12,3))
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()

Credit
timeseries for fastai2 was inspired by Ignacio's Oguiza timeseriesAI (https://github.com/timeseriesAI/timeseriesAI.git).> Inception Time model definition is a modified version of [Ignacio Oguiza] (https://github.com/timeseriesAI/timeseriesAI/blob/master/torchtimeseries/models/InceptionTime.py) and [Thomas Capelle] (https://github.com/tcapelle/TimeSeries_fastai/blob/master/inception.py) implementaions